EQL Projects
Tools to Track Sources of Fecal Pollution
Microbial source tracking (MST) is a tool that uses FIB that are unique to a specific host. With this tool, measurements of FIB can be tied to a specific source so that targeted remediation plans can be formed. Microbial source tracking can also be used to support the development and implementation of Total Maximum Daily Loads (TMDLs).
Tracking Fecal Pollution Links
Microbial source tracking needs to be used as part of a multi-tiered approach to water quality investigation.
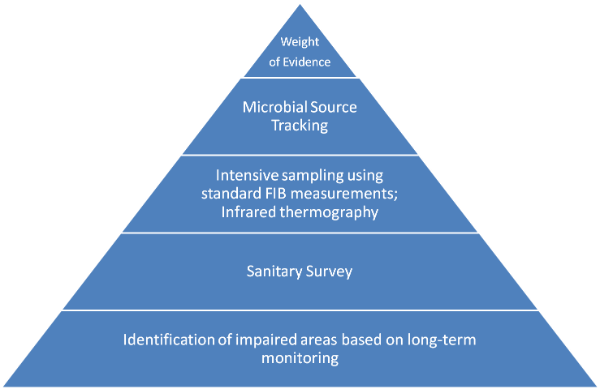
It is important to start at the bottom of the pyramid and move towards the top. In a project aiming to reduce FIB levels, two questions need to be answered:
- Where are we seeing the highest concentrations of FIB?
- What are the sources of FIB at sites where we are seeing the highest concentrations of FIB?
Microbial source tracking should be used to answer the second question, only after the first question has been addressed. Why? In order to use microbial source tracking most effectively, we need to draw on existing information to direct our MST efforts at those sites that are most contaminated.
Before Microbial Source Tracking: Identifying Sites with High FIB Concentrations
 The first step is to identify impaired areas based on data from long-term monitoring. Data collected as part of routine regulatory compliance can be used for this step. It is often useful to map the data in a Geographic Information System (GIS) to identify geographic hot spots of bacterial contamination. The second step is a sanitary survey. A sanitary survey is a detailed visual survey of the impaired area’s watershed for obvious sources of FIB such as piles of bird feces or leaking sewer lines.
The first step is to identify impaired areas based on data from long-term monitoring. Data collected as part of routine regulatory compliance can be used for this step. It is often useful to map the data in a Geographic Information System (GIS) to identify geographic hot spots of bacterial contamination. The second step is a sanitary survey. A sanitary survey is a detailed visual survey of the impaired area’s watershed for obvious sources of FIB such as piles of bird feces or leaking sewer lines.
Microbial Source Tracking to Identify FIB Sources
Third, more intensive sampling using standard FIB measurements should be conducted to gain a higher resolution picture of the FIB problem in space and time. Ideally, samples would be collected more frequently, at more sites, and during both dry and wet weather conditions. Other tools, such as infrared thermography can be used to supplement samples and gain information covering a larger geographic area. Once we have gained a solid understanding of the location of FIB-impaired areas, microbial source tracking can be used to determine the sources of FIB.
 Now, we are ready to use microbial source tracking. We mentioned above that microbial source tracking is a tool that uses FIB which are unique to a specific host, allowing FIB measurements to be tied to both a host and location. The process by which we measure these host-specific FIB is called quantitative Polymerase Chain Reaction (qPCR). qPCR allows us to isolate and identify FIB based on their DNA.
Now, we are ready to use microbial source tracking. We mentioned above that microbial source tracking is a tool that uses FIB which are unique to a specific host, allowing FIB measurements to be tied to both a host and location. The process by which we measure these host-specific FIB is called quantitative Polymerase Chain Reaction (qPCR). qPCR allows us to isolate and identify FIB based on their DNA.
It is important to note that microbial source tracking is often an iterative process. So, multiple rounds of testing may be needed to pinpoint sources to a specific host and location. qPCR assays (tests) for many different animals – including humans, dogs, birds, etc. – are already published in the scientific literature. We take these published assays and validate them for local use in a preliminary study since bacterial DNA can vary by geographic region. Once the assays are validated, we can then use them to determine the sources of FIB in that region.
Weight of Evidence Approach
No single tracer is a silver bullet because not all tracers move through the environment in the same way. So, if we apply a weight of evidence approach using multiple tracers, we can validate the results generated by our qPCR tracers with other tracers, garnering more confidence in our results. This means that we need to analyze each water sample for qPCR tracers, cultural eutrophication tracers, and chemical human tracers. Cultural eutrophication tracers and chemical human tracers are indicative of human waste and bacterial transport. It is also a good idea to continue measuring the regulated FIB that initially prompted the project so that the MST results can be framed in a regulatory context. Below you can see a partial list of tracers that could be used in a weight of evidence approach along with their purpose.
|
Tracer Type |
Tracer |
Purpose |
| qPCR Tracers | Bacteriodes thetaiotamicron (GenBac) | Identifies general mammalian sources |
Bacteriodes dorei (BacHum) |
Identifies human sources |
|
Bacteriodes canine (BacCan) |
Identifies canine sources |
|
| Regulated FIB | E. coli | Frames results in regulatory context |
Enterococcus |
Frames results in regulatory context |
|
| Cultural Eutrophication Tracers | Ammonia | Tracer of urine |
Biological oxygen demand |
Measures strength of oxygen-depleting constituents in water |
|
Turbidity |
Measures water’s murkiness |
|
Total suspended solids |
Measures solids content of water |
|
Volatile suspended solids |
Measures organic content in water |
|
| Chemical Human Tracers | Optical brighteners | Constituent in detergent–tracer for human sources |
Caffeine |
Passed through humans undigested–tracer for human sources |
How would this work? For example, if we see high concentrations of ammonia and caffeine in water that also has a strong BacHum signal, we can make a powerful argument for human sources of FIB in that watershed. Remediation efforts could then be focused on reducing human sources in that watershed.
Collaboration
The overarching goal in a microbial source tracking project is usually to get a water body in compliance with water quality regulations. To accomplish this goal, we cannnot overemphasize the importance of a close collaboration between stormwater managers and researchers.
We applied these tools in our pilot study basin, the Withers Swash basin in Myrtle Beach, South Carolina.